*The short version: at the end of the post there’s some fast code (Matlab with MEX) for training an HMM for spike sorting*
Earlier, we evaluated different solutions for sorting spikes for our 96-electrode array. One solution we looked at was spike sorting with HMMs (Hidden Markov Model). While most spike sorters first detect spikes and subsequently work on “snippets”, an HMM works directly with the full signal. The most appealing property of the HMM sorter is that it is highly sensitive; roughly speaking, the static detection filter is replaced by a filter adapted to the waveforms. The joint determination of the waveforms and their position is done through EM, specifically the Baum-Welch algorithm.
Joshua Herbst has an example implementation of Baum-Welch. It is meant to be readable, and is not optimized in any way. I wanted to find out how practical the HMM sorter would be with the large amounts of data generated by an electrode array. We can stress test the implementation by simply duplicating the example data:
data2 = repmat(data,1,250); tic;[mu1,sigmaz,pz] = hmm_Baum_Welch(data2,mu,sigma,p);toc;
On my computer, this takes about 110s and eats up 6 GB of RAM. Since the original data is 25000 samples long, then this is only 4 minutes of data at 25kHz, so this won’t work for the amount of data that we have. There’s two problems we must attack: memory usage and speed.
Memory usage can be brought down through a simple algorithmic change. In Baum Welch, and
(the forward and backward probabilities at time sample i) are computed recursively, and then multiplied together to obtain
and used to estimate various quantities of interest. These quantities of interest (typically means) can be computed incrementally, like so:
sumOfStuff = 0; For i = 1:numObservations ComputesumOfStuff +=
* somethingOrOther_i Discard
End For meanOfStuff = sumOfStuff/numObservations
The problem is that cannot be computed incrementally, because it requires
which is computed through a backwards recursion from the end of the data vector. What we can do is compute
in a first run and keep only every N’th value. These values are called checkpoints, and the resulting algorithm called the checkpoint algorithm. Once we have these checkpoints, we can compute
on the fly using the standard forward and backward algorithms. It goes like this:
sumOfStuff = 0; N = the block size; checkpoints = computebut only memorize every Nth value For j = 1:numObservations/N
= compute N forward iterations from latest
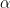
= compute N backwards iterations from checkpoint_j
For i = 1:numObservations sumOfStuff +=
* somethingOrOther_i End For Discard
End For meanOfStuff = sumOfStuff/numObservations
The maximum memory saving with the checkpoint algo is , where L is the number of observations, which is huge; for 6.25M observations, we’re talking 1200x times less memory. The CPU overhead for the extra beta run is only 1.5x, so overall this is very much worth it.
What about computation time? Baum-Welch involves a non-parallelizable for loop of death, the kind of stuff that Matlab chokes on. But it’s pretty straightforward to code this up in C and wrap as a MEX file (see also this). With this solution in hand the initial test lasts 14s with a blocksize of 500,000, and takes less than 700MB of RAM; with a blocksize of it takes 16s and less than 20MB of RAM (not a typo).
This plug-in replacement for Baum-Welch is exact (within machine precision). We might stop there, but we can shave off a bit more time with the observation that no neurons are firing at the vast majority of time points. Thus at most times, alpha is very close to [1,0,0,…], and we can modify the inner block iterations by setting the “latest ” to this guess, like so:
sumOfStuff = 0; N = the block size; checkpoints = compute beta but only memorize every Nth value startAlpha = [1,0,0,0,0...] For j = 1:numObservations/N in any order= compute N forward iterations from startAlpha
= compute N backwards iterations from checkpoint_j
For i = 1:numObservations sumOfStuff +=
* somethingOrOther_i End For Discard
End For meanOfStuff = sumOfStuff/numObservations
This might not sound like a big change, but it allows us to parallelize the outer for loop since the iterations are now independent of each other. How good is the approximation? Pretty good actually. On this example dataset the relative difference between the states means derived from the original and approximate methods is on the order of 1e-8 for a blocksize of about 100,000. Any inaccuracy in will decay very fast (the fact that we may have missed a spike will not matter for how we interpret the signal 10 ms afterwards). We just need to make sure that the blocks are long enough so that the potential number of spikes on the edges of the blocks is low relative to the total number of spikes. beta does not behave this way though so the initial checkpoint run is still required.
With this change and the use of parfor it now takes about 7s for a single iteration (15x faster than the original version, 2x faster than the serial version) on an i7 quad core and about 200MB of memory (parfor has quite a bit of memory overhead). You could probably bring this down more with some better optimized C code (I’m no expert in C). The parallel bit could potentially be done on the GPU, which could be even faster.
Point is, training an HMM for spike sorting is practical for long recordings, with a little work. I have not discussed determining the most likely path for the HMM, that will wait for another day, but I will say for now that you can replace Viterbi with a greedy algo as discussed in Sahani’s thesis and it works almost as well while being much less expensive. Update: I’ve received news from the Hahnloser lab that such an approximate alternative to Viterbi, as well as another training algorithm implemented in C are to be posted in the near future. Stay tuned.
Here are the files. It includes some code from J Herbst which is under the GPL and by extension the whole thing is GPL. You will need to have Matlab set up for compiling mex (I’m on GCC w/ 64-bit Linux). My implementation should support multiple neurons although I haven’t actually tested it. The mex files don’t do any sort of input checking and will probably crash Matlab if you call them with the wrong arguments. Use at your own risk.
3 responses to “Is it practical to train an HMM for spike sorting with Baum-Welch?”
The files are missing again… Could you repost them, please?
Do you think you could re-post your files, I’m interested in playing around with this a bit, and there is little to no other info out there on this that I can find.
Yes, updated.